Yang Laboratory |

|
Structures and Functions of DNA G-quadruplexes
DNA G-quadruplexes (G4) are novel non-canonical DNA secondary structures that form in biologically important Guanine-rich regions in human genome. G-quadruplexes can readily form under physiological conditions and exhibit great structural diversity. We seek to understand the molecular structures and functions of the biologically relevant DNA G-quadruplexes, including those formed in the proximal promoter regions of human oncogenes as well as those formed in human telomeres. G-quadruplexes formed in the promoters of human oncogene, such as c-MYC, BCL-2, VEGF, and PDGFR-b, provide a potential means to modulate oncogene transcription. G-quadruplexes formed in the human telomeres can inhibit the cancer-specific enzyme telomerase and telomere maintenance. G-quadruplexes have become one of the most exciting nucleic acid secondary structures. Delineating the molecular level details of the biologically relevant G-quadruplexes and their drug and protein complexes is important for the evaluation of their potential as cancer therapeutic targets as well as for structure-based rational drug design.

G4-targeting Drug Discovery
The biologically relevant DNA G-quadruplexes, in particular those formed in human oncogene promoters and telomere, are emerging as promising new molecular targets for cancer therapeutics. We seek to discover and develop small molecular anticancer drugs that target the DNA G-quadruplexes using high-throughput screening in combination with biochemical, biophysical and cellular methods, as well as structure-based rational design. We work to delineate the molecular level details of small molecule interactions with biologically relevant G-quadruplexes, to understand the mechanisms of action of small molecules, and to conduct structure-based rational design of G4-targeted drugs.
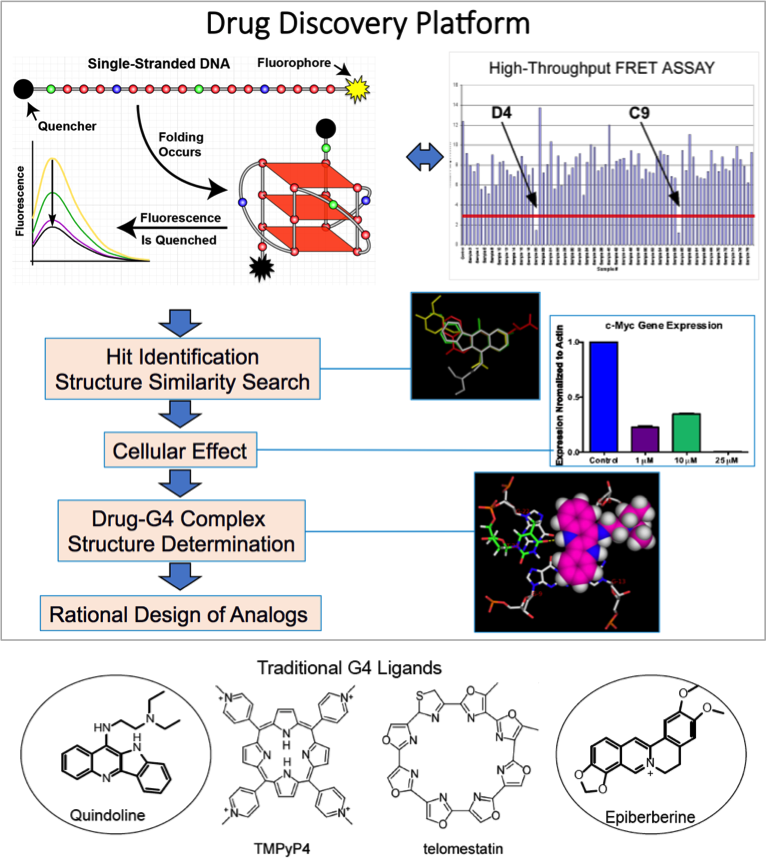
Target the MYC promoter G4 to suppress MYC oncogene
The identification of globular G4 DNA structures in genomic regions regulating gene expression has created the opportunity to selectively target G4s as a new class of drug receptor. The transcription factor MYC is one of the most important oncogenes deregulated in the majority of cancers, however, the MYC protein is not an easy drug target. We aim to target the MYC oncogene through the MYC promoter G4. We work to study the structural basis of small molecule recognition of MYC G4 and carry out structure-based rational design of MycG4-targeting small molecule drugs.
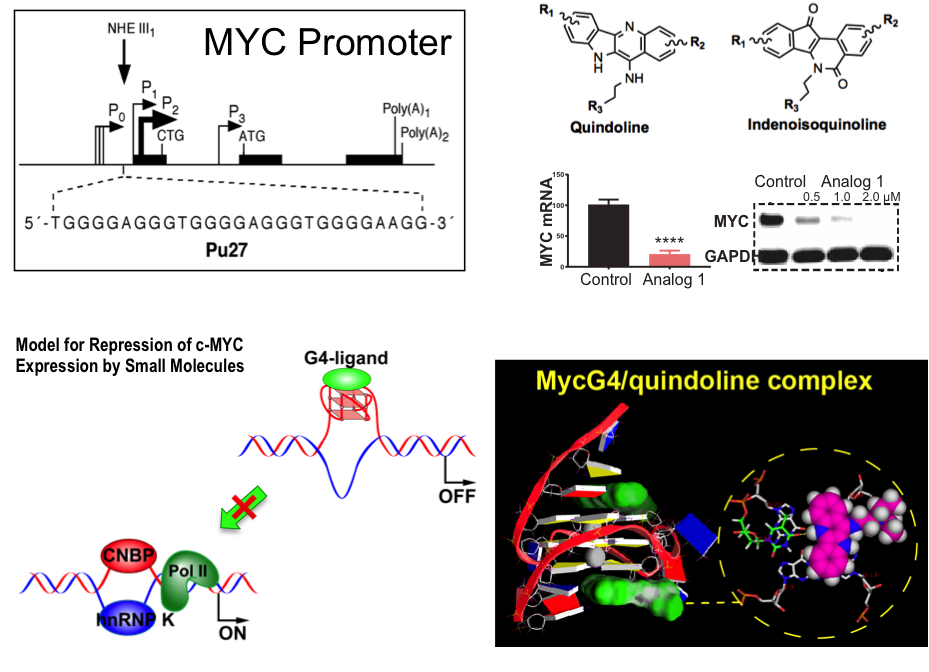
Ambrus et al., Biochem, 2005; Dai et al., JACS, 2011; Wang et al., JACS, 2019.
Targeting Human Telomeres
Human telomeres play critical roles in cancer, aging, and genetic stability. The 3′-overhang of human telomeres can form DNA G-quadruplex (G4), an attractive target for anticancer drugs. Small molecules that stabilize the telomeric G4 can inhibit cancer-specific enzyme telomerase, disrupt telomere maintenance and result in cancer cell apoptosis. We work to understand the molecular basis for specific recognition of the hybrid-2 (major) and hybrid-1 (minor) telomeric G4 formed in physiologically relevant K+ solution. We hope to elucidate the structural basis for rational drug design targeting the human telomeric G4.
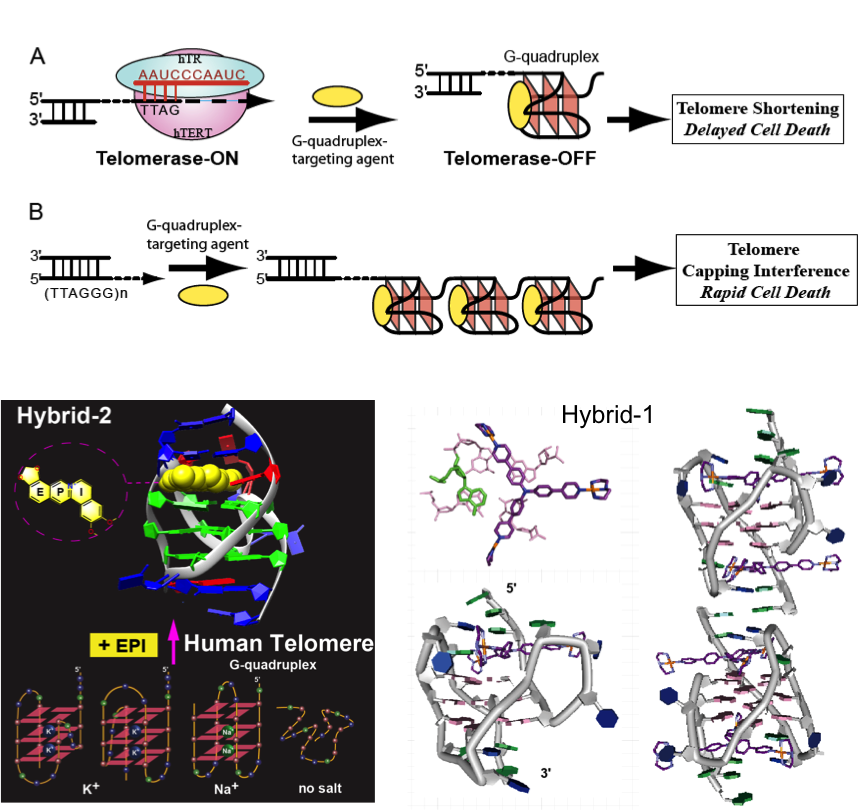
Ambrus et al., NAR, 2006; Dai et al., NAR 2007; Lin et al., Angew. Chem. 2018; Liu et al., Nature Comm, 2018
Protein interactions of DNA G-quadruplexes
G4-DNA-interactive proteins are important for the cellular functions of G4 DNA. Particularly, proteins interact with the c-MYC promoter G-quadruplex are involved in c-MYC gene regulation. Nucleolin specifically binds and stabilizes the c-MYC G-quadruplex and functions as a transcription repressor. We have recently discovered DDX5 as a new G4 resolvase/helicase, which unfolds the c-MYC G-quadruplex and functions as a transcription activator. We work to understand the molecular level interactions of these proteins with the c-MYC G-quadruplex, their cellular functions in relation to chromatin structure, and their potential for small molecule targeting.
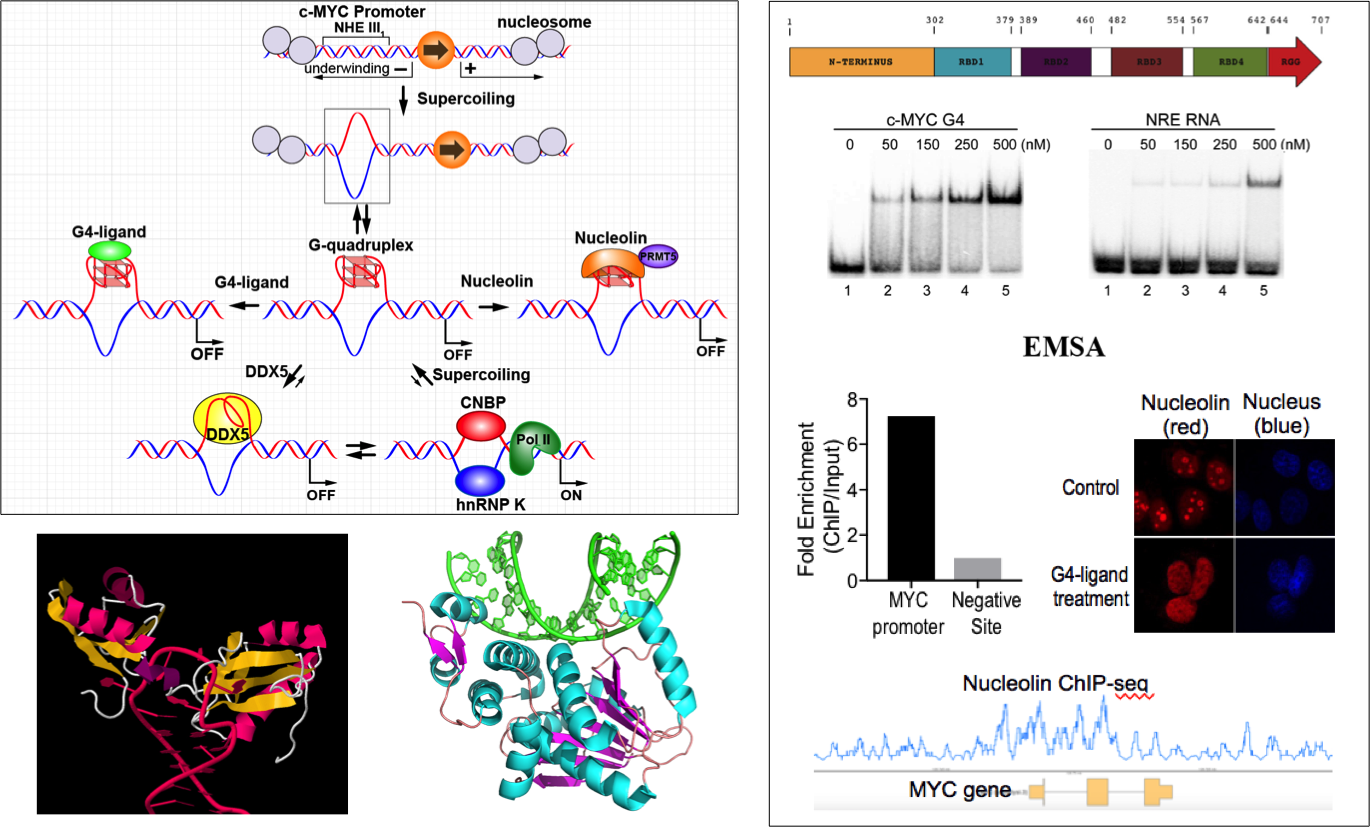
Wu et al., PNAS, 2019